19 November 2020
Researchers from CSIRO, Charles Darwin University and The University of Western Australia have developed a machine-learning approach that reliably detects invasive gamba grass from high-resolution satellite imagery.
Gamba grass, originally from Africa, is listed as a Weed of National Significance, and is one of five introduced grass species that pose extensive and significant threats to Australia’s biodiversity. The perennial grass can grow to four metres in height and forms dense tussocks which can burn as large, hot fires late in the dry season.
Mapping where gamba grass occurs is essential to managing it effectively but northern Australia is so vast and remote that on-the-ground mapping and even airborne detection of the weed is too labour-intensive. So the researchers turned to high-quality satellite imagery and developed a technique that could help detect and prioritise gamba grass for management.

Gamba can grow up to four metres in height and form dense tussocks. Photo by NESP Northern Hub.
Dr Shaun Levick from Australia’s national science agency, CSIRO, said that the research team used field data to ‘train’ a machine-learning model to detect gamba grass from high-resolution, multispectral satellite imagery.
Under optimum conditions, our method can detect gamba grass presence with about 90 per cent accuracy.
– Dr Shaun Levick, CSIRO
The researchers commissioned the WorldView-3 satellite to capture very high-resolution imagery across 16 spectral bands for an area of 205 square kilometres near Batchelor in the Northern Territory – an area of dense gamba grass infestation. The wide range of spectral data allowed them to use factors unseen to the human eye, such as leaf moisture levels and chlorophyll content, to differentiate between gamba grass and native grass species.
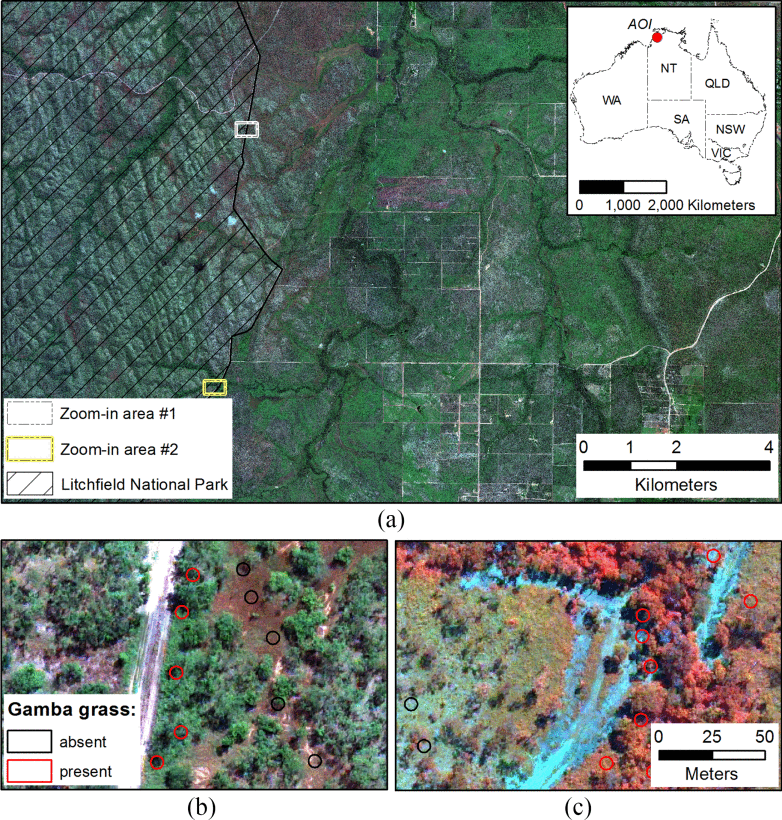
WorldView-3 image of (a) Area of interest in relation to the extent of Australia with a (b) zoom-in area #1 in true-color and (c) zoom-in area #2 in false-color. 10.1109/JSTARS.2020.3013663
Dr Natalie Rossiter-Rachor, of Charles Darwin University, said that the project drew on extensive on-ground research into the life cycle of gamba grass to help achieve such accurate detection rates.
We knew that gamba grass tends to stay green longer into the dry season than native grasses, so we timed the capture of the satellite imagery for this period. Understanding the ecology of the problem was essential to informing the remote sensing and machine-learning solution to the problem.
– Dr Natalie Rossiter-Rachor, Charles Darwin University
The project, funded by the Australian Government’s National Environmental Science Program under the Northern Australia Environmental Resources Hub, is part of a larger effort to detect and map gamba grass throughout the north.
Our longer-term goal is to move to a system where we can use free, open-access imagery to map gamba grass. We want to develop a technique that is accessible to anyone and that can help improve land management in northern Australia.
– Dr Shaun Levick
Associate Professor Samantha Setterfield from The University of Western Australia said that accurate maps of where gamba grass occurs are essential to control the spread of the weed.
Mapping gamba grass using satellite imagery unlocks the potential to frequently map large areas so we can get a better picture of where gamba grass is across northern Australia, and how quickly it is spreading. Managers can then target areas that are the highest priority for control, such as biodiversity-rich areas or culturally important sites.
– Associate Professor Samantha Setterfield, The University of Western Australia
Dr Shaun Levick at CSIRO led this study. Photo: NESP Northern Hub.
Want to know more about the Resilient Landscapes Hub's activities and our research into practical solutions to environmental problems? Stay informed about activities, research, publications, events and more through the Hub newsletter.